Part:BBa_I766555:Experience
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_I766555
User Reviews
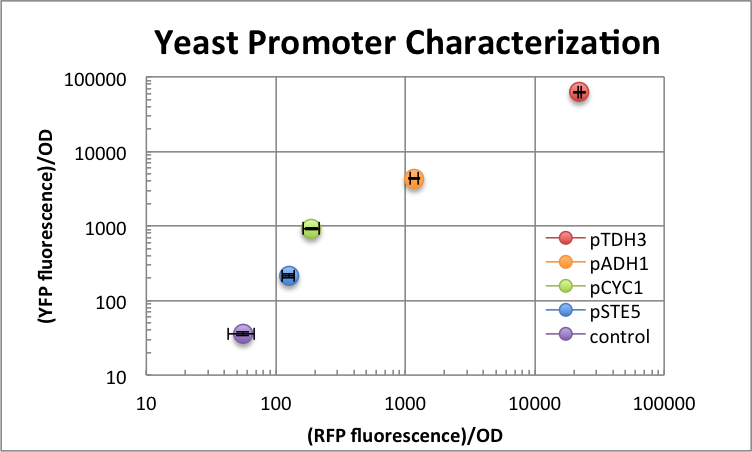
[http://2012.igem.org/Team:Berkeley/Project/Localization#Promoter%20CharacterizationUCBerkeley 2012 Team:] To assay their potential use in our project, we characterized the strength of several registry yeast promoters. We wanted to compare their consistency of expression and relative fluorescence, then quantify them against a standard promoter fluorescence. The promoters we used in our assay: pSTE5 (weak), pCYC1 (medium),pADH1 (strong), and pTDH3 (strongest).
Experimental Design: We designed our promoters to express a fluorescent protein so that we could quickly measure bulk fluorescence via TECAN. To test if downstream sequence affected expression, we cloned the promoter in front of two different fluorescent proteins, yellow fluorescent Venus and red fluorescent mKate in different strains. If expression was sequence-independent, we expected similar fluorescence intensity from both channels. The device was cloned on a backbone with Leu2 marker and Cen6 origin of replication and transformed into S228C S. cerevisiae. Ideally, we wanted the calibration curve to span several orders of magnitude and exhibit a linear relationship, which was true for all the promoters we characterized except for pCYC1.
| Promoter | RFP/OD | YFP/OD |
| background | 55±10 | 36±2 |
| pSTE5 | 126±10 | 216±20 |
| pCYC1 | 189±30 | 928±30 |
| pADH1 | 1170±90 | 4360±100 |
| pTDH3 | 22164±700 | 62531±2000 |
UNIQ7b936620a931d729-partinfo-00000000-QINU UNIQ7b936620a931d729-partinfo-00000001-QINU